A complete cloud solution for bioinformaticians will be available at the Center for Biotechnology (CeBiTec) at Bielefeld University, including computing and storage infrastructure (IaaS), a framework for simplified access to the cloud infrastructure (PaaS) and ready to use bioinformatic workflow solutions (SaaS) in the field of metagenomics. The provided cloud environment involves the following services:
- Infrastructure as a Service (IaaS): highly advanced users are in full command of the tools to build workflows and pipelines that require performant infrastructure for data analysis. The users access the infrastructure via the virtualization system, such as virtual machines or Docker containers.
- Platform as a Service (PaaS): We provide BiBiGrid as a tool for easy HPC cluster setup inside a cloud environment. It simplifies the often complex configuration and setup of typical software stacks to a single command line call.
- Software as a Service (SaaS): One major goal of the de.NBI cloud site in Bielefeld is to make popular bioinformatics workflows available to cloud users.
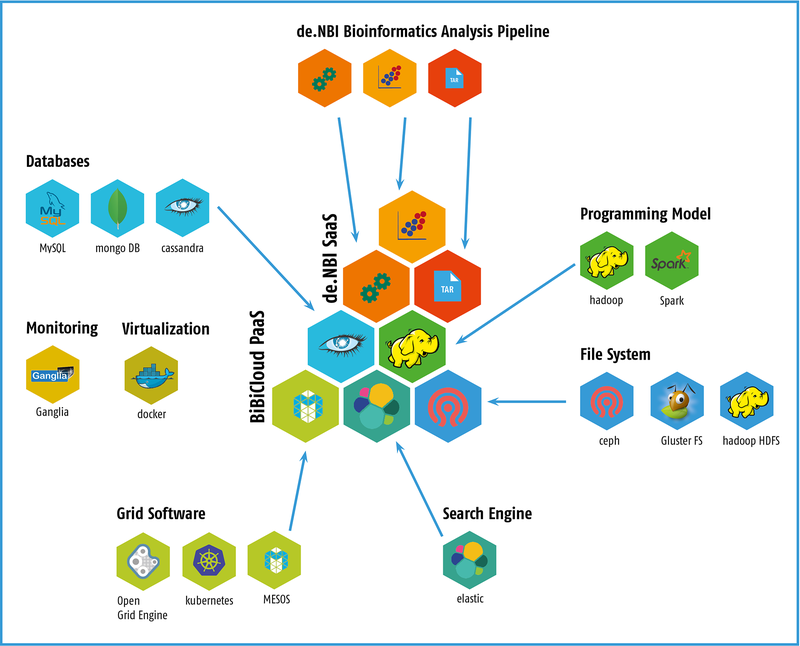
Popular bioinformatics tools and pipelines will be made available by pre-configured VMs or Docker containers. The focus of the Bielefeld Service Center is microbial genomics and metagenomics, as well as postgenomics applications. Within the de.NBI cloud, data sets and preconfigured pipelines for analyses in these research areas will be provided to a wide audience. This includes in the area of metagenomics 16S rRNA based as well as WGS-based analysis workflows, including large-scale assemblies and functional annotation. For these special applications, VMs with up to 2TB of RAM can be provided.
A special focus is also the analysis of Oxford Nanopore sequence data and postgenomics applications, where we e.g. provide resources for the MetaProteomeAnalyzer Service (MetaProtServ).
The de.NBI cloud infrastructure at Bielefeld University comprises beside general purpose resources, high memory instances with local disk space (up to 2 TB RAM and 5 TB local disk space) for applications such as metagenomic assemblies. For hardware accelerated machine learning applications special VMs with access to GPUs are available.