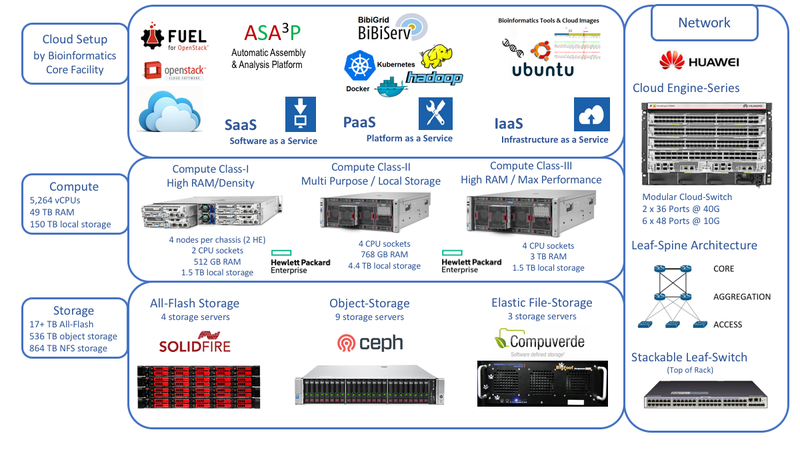
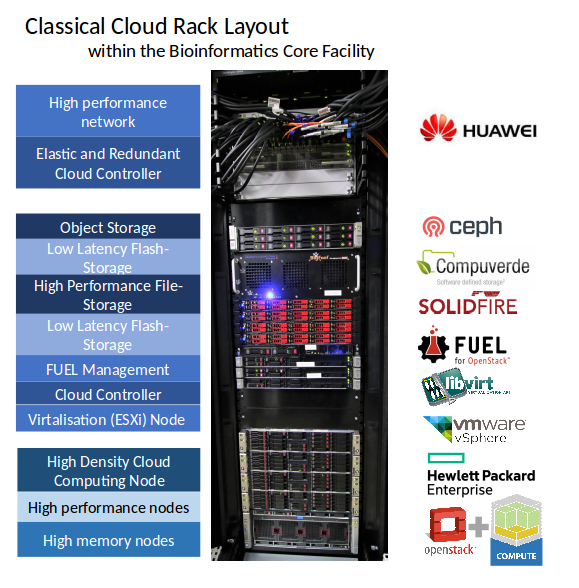
The de.NBI cloud infrastructure at Justus-Liebig-University (JLU) Giessen was successfully established in early 2017, providing large-scale computing and storage resources to users of the de.NBI network. An extension of the overall storage capacity is already underway and will be available in the first half of 2018.
After initial setup and intensive testing, the cloud at JLU Giessen was transferred to production mode in June 2017. It has already been used successfully to conduct practical sessions during the first de.NBI summer school on cloud computing for bioinformatics. On this occasion, 24 participants applied cloud computing techniques for the analysis of their own biological data sets and learned how to implement scalable bioinformatics software solutions in a cloud environment. Since its establishment, the cloud is regularly used by various cooperation partners and users of the de.NBI network to analyze their data.
With between 6 and 27 GB of main memory available for each CPU core, the generous equipment of the de.NBI cloud at JLU Giessen provides an ideal resource to execute tools and workflows with high memory requirements such as the assembly of large-scale metagenomes or eukaryotic genomes. Instance types with up to 3 TB of main memory and over 100 CPU cores are offered for this purpose.
The computing resources for running virtual machines are complemented by a selection of various storage solutions, including SSD-based ephemeral disks, data volumes and object storage, which can be used to store raw and intermediate data as well as the final results of bioinformatics analysis pipelines.
The de.NBI cloud team at JLU Giessen provides ready-to-use images for microbial data analysis, assembly and statistical processing. Also, we will soon offer various preselected and readily usable biological databases and sequence collections like NCBI GenBank, RefSeq, Pfam or preprocessed human genomes, which will be made available via a shared file system for immediate use. Thus, users can start analyzing their data right away without the tedious need to collect and preprocess required databases.
In cooperation with our BiGi partners at Bielefeld University, the BiBiGrid framework is available, which provides an easy means to setup cloud-based computing clusters to process large data amounts in parallel, as e.g. required for processing metagenome or transcriptome data. On top of the BiBiGrid framework, we provide one of the most recent additions to our portfolio of bioinformatics applications, the ASAP application, which can be used for the concurrent analysis, assembly, annotation and comparative analysis of bacterial genomes. Based on this unique combination and powered by the de.NBI cloud at JLU Giessen, we are currently able to easily analyze thousands of microbial genomes each day.
In addition to the general-purpose computing servers, we have several ActiveMotif Decypher FPGA-based systems available, which provide hardware-accelerated versions of selected bioinformatics applications, such as sequence homology searches required for genome analysis and annotation as well as metagenome data processing. Currently, we are evaluating several solutions to make these systems also available to our cloud users. Furthermore, simplified deployment of Hadoop-based elastic and flexible data analysis environments, customizable by each individual user, will be offered in the first half of 2018.
As of summer 2017, the de.NBI cloud setup in Giessen spans about 80 hosts with more than 2,600 cores, a total of 48 TB RAM, 140 TB of local SSD storage in compute hosts, and about 500 TB of distributed storage for volumes and object storage.