The Heidelberg Center of Human Bioinformatics HD-HuB, provides a cloud solution for life scientists, including rich data sets and standardized pipelines as well as bare compute infrastructure in order to meet the wide range of user requirements spanning the biological research community.
Together the HD-HuB partner institutions have experience in working with a broad range of biological datasets, developing state-of-the-art bioinformatics methods and infrastructure for their analysis. Our particularly strong expertise in processing large-scale human genomic and imaging data (e.g. PCAWG, ICGC) influences the Heidelberg de.NBI cloud’s major focus on data and infrastructure. We aim at the standardization of analysis approaches to improve comparability of data sets across institutions, to enable integrative analyses and meta-analyses. We are committed to the FAIR principles of data sharing.
The major goal of the de.NBI cloud site in Heidelberg is to make state-of-the-art tools for analyzing human genomics, metagenomics, and microscopy image data widely accessible.
Services are provided in all of the „classic“ cloud areas:
- Infrastructure as a Service (IaaS): Users are in full control of their virtual infrastructure. Resources are provided not only in the form of compute via virtual machines (VMs) but also include networks, firewalls and storage. Auxiliary, we offer specialized hardware (e.g., GPUs, FPGAs and high-memory nodes) for state of the art large scale parallel scientific computing (e.g., Matrix Factorization, Deep Learning) to the community. This gives users the flexibility they need to match hardware to their requirements and efficiently develop workflows and pipelines, while optimizing for performance.
- Platform as a Service (PaaS): Users are provided a „platform“ that allows them to accomplish their scientific tasks. These are usually provided in the form of workflow frameworks such as Galaxy https://github.com/BMCV/galaxy-image-analysis, Roddy, Butler (which features multi-cloud deployment, configuration management, comprehensive monitoring, and anomaly detection as well as pre-built pipelines for genome alignment, germline and somatic variant calling, and R script execution) or the ICGC pipelines but are not limited to that; we aim to provide several VM images, each with a curated set of software focused on a specific area of bioinformatics, to allow life science researchers to get started quickly.
- Software as a Service (SaaS): Users are provided fully established software packages or workflow systems that can be directly used and applied to their problems and data. This is ideally suited for exploratory analyses of genomics and metagenomics, imaging data and beyond. Additionally, we provide customized software and workflows in the context of sharing our expertise to cooperation partners within the de.NBI community. It is also a great resource for novice bioinformaticians and to aid teaching.
- Data as a Service (DaaS): Users are provided access to large data pools that are essential to life science research. Those are a highly valuable resource but often cannot be transferred or stored at the users‘ home organization due to the size or access regulations of the data. By enabling access to these data the de.NBI cloud will drive the development of new bioinformatics methods, and facilitate benchmarking of analysis techniques on previously inaccessible datasets. Depending on their need, users can be granted authorized access to restricted data sets such as reference data sets from the EBI-ENA or common data sources such as dbGaP and GENCODE. We started to join the new ICGC cloud computing project for collaborative research. The raw and interpreted data processed from the sequences of tumours and matching normal tissues will be provided by the Heidelberg de.NBI Cloud in order to generate analytic results and to develop new tools.
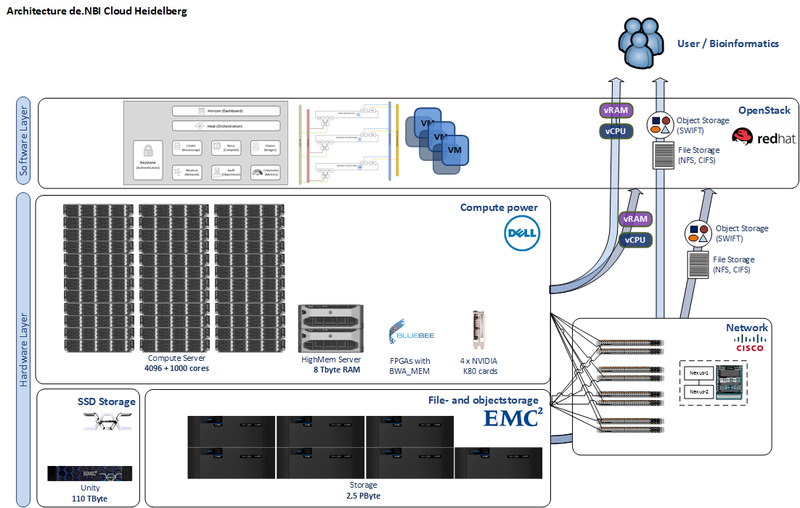
The de.NBI cloud infrastructure in Heidelberg is comprised of 4096 cores, 32 TByte RAM, 8 TByte RAM in high-memory nodes, 100 TByte SSD and 2,5 PByte storage capacity. FPGAs ensure rapid processing of genomic alignments. GPUs are available for deep learning approaches on massive amounts of labeled data.